-Search query
-Search result
Showing all 22 items for (author: nicholas & rj)

EMDB-25660: 
Cryo-EM structure of Csy-AcrIF24
Method: single particle / : Mukherjee IA, Chang L
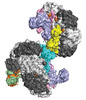
EMDB-25661: 
Cryo-EM structure of Csy-AcrIF24 dimer
Method: single particle / : Mukherjee IA, Chang L

EMDB-25662: 
Cryo-EM structure of Csy-AcrIF24-DNA dimer
Method: single particle / : Mukherjee IA, Chang L

EMDB-25788: 
Cryo-EM structure of the Csy-AcrIF24-promoter DNA dimer
Method: single particle / : Mukherjee IA, Chang L

EMDB-25789: 
Cryo-EM structure of the Csy-AcrIF24-promoter DNA complex
Method: single particle / : Mukherjee IA, Chang L

PDB-7t3j: 
Cryo-EM structure of Csy-AcrIF24
Method: single particle / : Mukherjee IA, Chang L

PDB-7t3k: 
Cryo-EM structure of Csy-AcrIF24 dimer
Method: single particle / : Mukherjee IA, Chang L

PDB-7t3l: 
Cryo-EM structure of Csy-AcrIF24-DNA dimer
Method: single particle / : Mukherjee IA, Chang L

PDB-7taw: 
Cryo-EM structure of the Csy-AcrIF24-promoter DNA dimer
Method: single particle / : Mukherjee IA, Chang L

PDB-7tax: 
Cryo-EM structure of the Csy-AcrIF24-promoter DNA complex
Method: single particle / : Mukherjee IA, Chang L

EMDB-23927: 
Cryo-EM structure of affinity captured human p97 hexamer
Method: single particle / : Hoq MR, Vago FS, Li K, Kovaliov M, Nicholas RJ, Huryn DM, Wipf P, Jiang W, Thompson DH

EMDB-23928: 
Cryo-EM structure of affinity captured human p97 double-hexamer
Method: single particle / : Hoq MR, Vago FS, Li K, Kovaliov M, Nicholas RJ, Huryn DM, Wipf P, Jiang W, Thompson DH

EMDB-22078: 
Characterization of the SARS-CoV-2 S Protein: Biophysical, Biochemical, Structural, and Antigenic Analysis
Method: single particle / : Herrera NG, Morano NC, Celikgil A, Georgiev GI, Malonis R, Lee JH, Tong K, Vergnolle O, Massimi A, Yen LY, Noble AJ, Kopylov M, Bonanno JB, Garrett-Thompson SC, Hayes DB, Brenowitz M, Garforth SJ, Eng ET, Lai JR, Almo SC

EMDB-4598: 
Rhodopsin-Gi protein complex
Method: single particle / : Tsai CJ, Marino J, Adaixo RJ, Pamula F, Muehle J, Maeda S, Flock T, Taylor NMI, Mohammed I, Matile H, Dawson RJP, Deupi X, Stahlberg H, Schertler GFX

EMDB-4483: 
Correlative FM and ET of GFP-Bax in HeLa cells
Method: electron tomography / : Ader NR, Hoffmann PC, Ganeva I, Borgeaud AC, Wang C, Youle RJ, Kukulski W

EMDB-4484: 
Correlative FM and ET of GFP-Bax in HCT116 cells
Method: electron tomography / : Ader NR, Hoffmann PC, Ganeva I, Borgeaud AC, Wang C, Youle RJ, Kukulski W

EMDB-4486: 
Correlative FM and cryo-ET of GFP-Bax in HeLa cells
Method: electron tomography / : Ader NR, Hoffmann PC, Ganeva I, Borgeaud AC, Wang C, Youle RJ, Kukulski W

EMDB-4490: 
cryo-ET of cryo-FIB milled HeLa cells overexpressing GFP-Bax
Method: electron tomography / : Ader NR, Hoffmann PC, Ganeva I, Borgeaud AC, Wang C, Youle RJ, Kukulski W

EMDB-4491: 
cryo-ET of cryo-FIB milled HeLa cell
Method: electron tomography / : Ader NR, Hoffmann PC, Ganeva I, Borgeaud AC, Wang C, Youle RJ, Kukulski W

EMDB-4492: 
cryo-ET of cryo-FIB milled Bax/Bak double knockout HCT116 cells stably expressing GFP-Bax
Method: electron tomography / : Ader NR, Hoffmann PC, Ganeva I, Borgeaud AC, Wang C, Youle RJ, Kukulski W
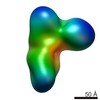
EMDB-8235: 
Negative stain structure of Vps15/Vps34 complex
Method: single particle / : Kirsten ML, Zhang L, Ohashi Y, Perisic O, Williams RL, Sachse C

PDB-5kc2: 
Negative stain structure of Vps15/Vps34 complex
Method: single particle / : Kirsten ML, Zhang L, Ohashi Y, Perisic O, Williams RL, Sachse C
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
